General-purpose LLMs like LLaMA and GPT-4 have demonstrated remarkable proficiency in understanding and generating natural language. However, their capabilities wane in highly specialized domains such as processing amino acid sequences for proteins (e.g., MTVPDRSEIAG) or SMILES strings for chemical compounds, hampering their adoption for a wide range of scientific problems. Further, Training domain specific LLM requires a significant amount of compute and in-domain data. Fine-tune existing LLMs or perform in-context learning might be another way to go around, but the former is prone to catastrophic forgetting and can hurt the model’s reasoning abilities
So can we effectively repurpose general-purpose LLMs for specialized tasks without compromising their linguistic and reasoning capabilities?
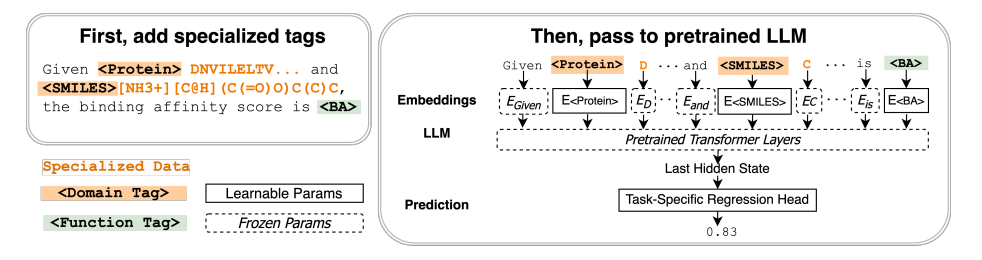
To address this issue, the paper has introduced Tag-LLM. A novel model-agnostic framework for learning custom input tags, which are parameterized as continuous vectors appended to the LLM’s embedding layer to condition the LLM. Two types of input tags: domain tags are used to delimit specialized representations (e.g., chemical formulas) and provide domain-relevant context; function tags are used to represent specific functions (e.g., predicting molecular properties) and compressing function-solving instructions. Further, a three-stage protocol is also been introduce to learn these tags using auxiliary data and domain knowledge.
Let’s consider the task of protein-drug binding affinity prediction. In Tag-LLM method domain tags ⟨Protein⟩, ⟨SMILES⟩ and a function tag ⟨Binding Affinity⟩ is been injects to the input, which are mapped to specially trained embeddings. The model’s last hidden state is passed to a task-specific head to generate predictions of the desired type (e.g., a scalar binding affinity value in this case).
While experimenting with the LLaMA-7B model, on a diverse set of ten domains, encompassing eight languages and two specialized scientific domains (protein sequences and SMILES molecule representations). Results demonstrate that the domain tags can act as effective context switchers, and a function tag can be applied to multiple domains to solve different tasks, achieving zero-shot generalization to unseen problems.